About the OH SFS Handbook¶
The work found in these pages got its start in the ORION project and continued in the frame of the BeOne project. The ORION project, launched in 2018, is aimed at establishing and strengthening inter-institutional collaboration and transdisciplinary knowledge transfer in the area of surveillance data integration and interpretation, along the One Health (OH) objective of improving health and well-being. The BeOne project, launched in 2020, aims at developing an integrated surveillance dashboard in which molecular and epidemiological data for foodborne pathogens can be interactively analysed, visualised, and interpreted by the relevant experts across disciplines and sectors.
Through three main work packages (WP), ORION’s specific goals can be summarized as the delivery of three main resources:
- a “OH Surveillance Codex” (WP1) - a high level framework for harmonised, cross-sectional description and categorisation of surveillance data covering all surveillance phases and all knowledge types;
- a “OHS Knowledge Hub” (WP2) - a cross-domain inventory of currently available data sources, methods / algorithms / tools, that support OH surveillance data generation, data analysis, modelling and decision support;
- “OHS Infrastructural Resources” (WP3) – that are practical, infrastructural resources forming the basis for successful harmonisation and integration of surveillance data and methods.
Through its WP1, the BeOne project targets the typing and nomenclature issues that exist within WGS-based pathogen surveillance and outbreak detection. The goals of this WP can be summarized as:
- establishing the current state of the art within genomics methods for WGS‐based typing;
- providing a strain dataset to capture the genomic diversity within the populations of four main pathogens: Salmonella enterica, Escherichia coli (STEC), Listeria monocytogenes and Campylobacter jejuni;
- assessing cluster agreement between different WGS-based typing approaches to ensure the comparability between distinct methodologies to reinforce and promote a global surveillance and control of infectious diseases.
The work in these pages springs out ORION’s WP2 and BeOne’s WP1 and focuses on being an inventory over current practices regarding the use of sequencing data for surveillance purposes, with a special focus on the methodologies used foodborne diseases and the current state of One Health surveillance.
Updates and contributions¶
The underlying technologies within this field is a moving target. It is thus important to keep this handbook updated with new information. Contributions to this handbook are very welcome, please see the Contributing document for more information.
Overview of the handbook¶
This handbook consists of various sections.
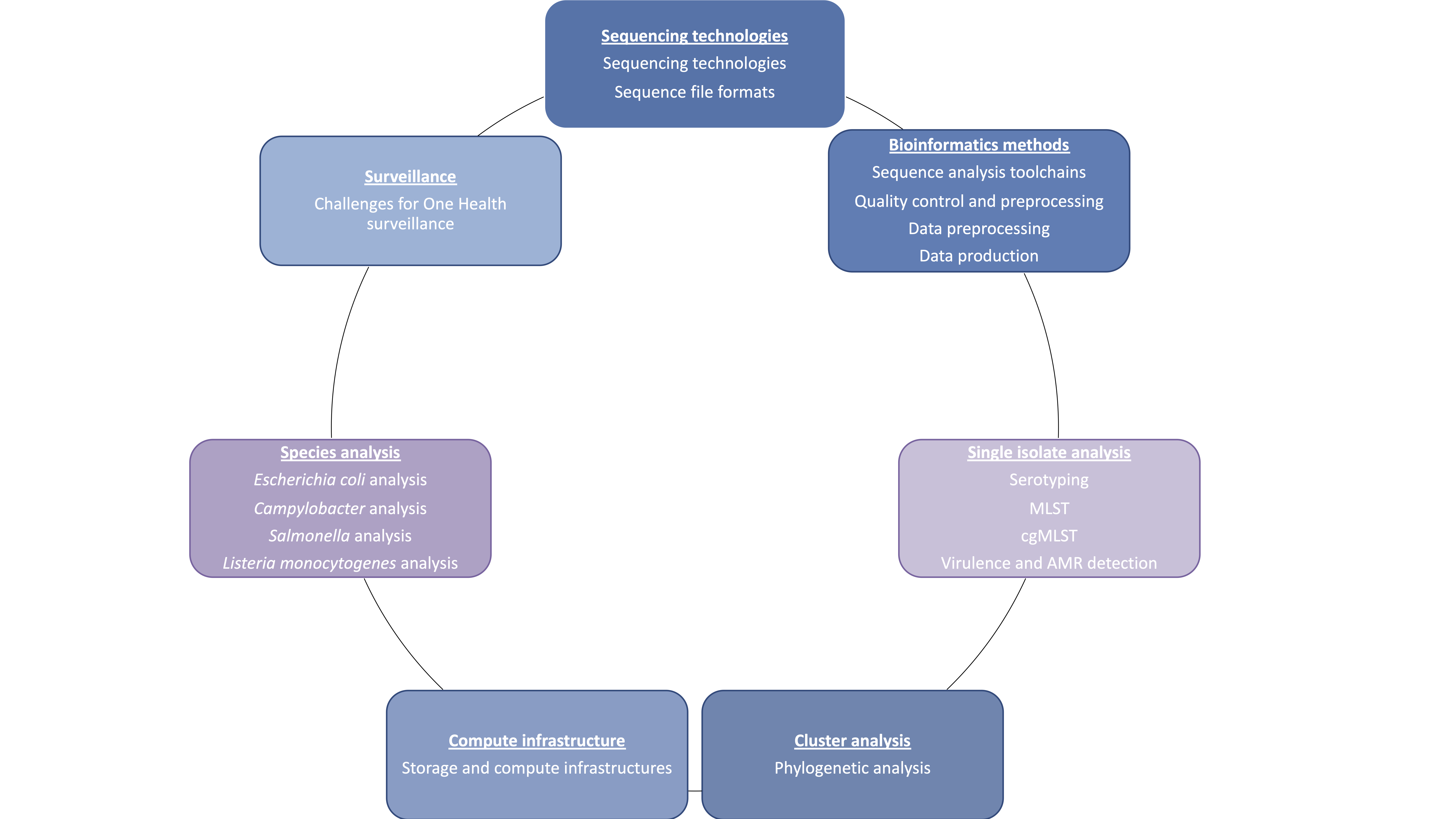
Sections of the handbook - Blue boxes describe species-agnostic processes, while the purple ones depend on which biological agent is being analysed.
Sequencing Technologies - The focus for this section is on describing the available sequencing technologies, highlighting their differences and consequent impact on WGS data analysis.
Bioinformatics methods - This section aims to describe the basic bioinformatics methods and tools that can be used to analyze whole genome seqencing data. This includes how to do quality control, how assembly and mapping works, and how a bioinformatics pipeline might be stitched together.
Single isolate analysis - This section explores different bacterial typing pipelines, including cg/wgMLST and SNP-based pipelines, and pipelines for virulence and antimicrobial resistance (AMR) detection.
Cluster analysis - The focus for this component is to provide an overview of the different methods which can be used to perform an integrated analysis of several samples, obtaining clustering information.
Compute infrastructure - The focus for this section is on exploring the options and the requirements for establishing possible infrastructures for using NGS methods for surveillance purposes. This section spans from storage, compute infrastructures and data management to workflow managers and currently available platforms for automated analysis.
Species analysis - This section is focused on four bacterial pathogens: S. enterica, E. coli (STEC), L. monocytogenes and C. jejuni, providing a historical overview of their respective typing methods, and exploring their specific needs and available pipelines/platforms for WGS surveillance. Their respective state of the art regarding WGS and One Health surveillance is also reviewed.
Surveillance - This section explores the relevance of WGS for One Health surveillance, and the challenges that we are currently facing for the implementation of an international and inter-sectoral surveillance.